Thanks @koerrie good catch! It’s fixed now. I actually had it right like it is now in my original code, but I guess that’s what I get for trying to simplify code I haven’t looked at in while 
You can just look at the corresponding object in the models list to find the parameters that go with any of the scores
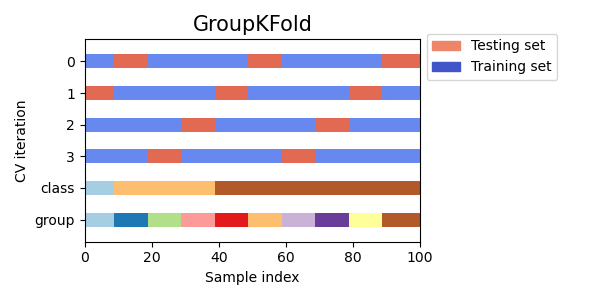
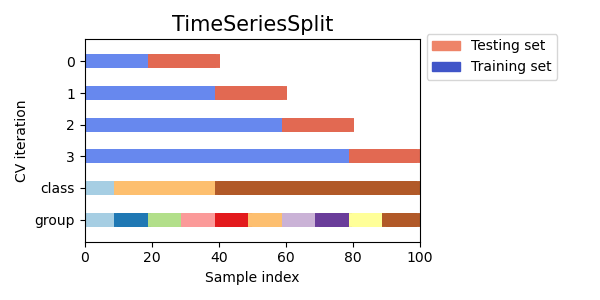
This is like the latter but splits are only between consecutive eras/groups which is not true for the sklearn version.
I’m fairly new to this so sorry if this is a dumb question. Once you find the optimal set of parameters, and you’re ready to fit the model, is there a way to incorporate the era groups? Or would you just use fit() as normal?
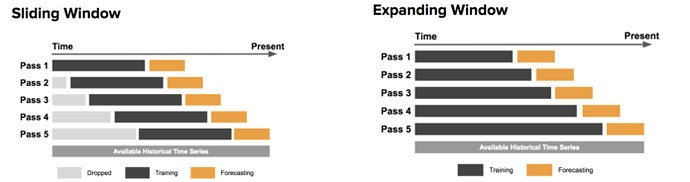
@mdo thanks for the original post and for the plot, really illustrative. I’ve been participating in Numerai for a while but also been working as a data scientist in time series problems. I’ve always used the TimeSeriesSplit approach to force testing on “future” data but always tried to keep using the same amount of training data in each fold.
I mean, if you are using 4 eras for training and 2 for testing in iteration 0, I’d prefer to use 4+2 also in iteration 1 and so on, but seems you’re using 8+2. I don’t have a strong opinion here, and since you are using the same approach for all models it should be ok. In my mind, it resonates as the average error metric won’t be accurate since I’d expect iteration 3 to be better (or worse if your model gets easily overfitted) than iteration 0 because it’s using more training data.
Edit: I’ve found an illustrative image for both approaches.
Is there some reasoning to use “Expanding window” over “sliding window”?
Many of my base models use time-series CV. Until @mdo made that post I did not think anybody else used it. For the models that I have using time-series-CV, I also used an expanding window. My reasoning is that I want the fold size to approach the size of the final training data set because if you use a fixed size which is much smaller than the final training data set size then you might train models that are too greedy and so there is a much greater chance of over-fitting. When we were only allowed 3 models I weighted those ensembles much more to the time-series CVed models and they reached the top 100 very quickly.
Edit: I clearly fail at reading and in-line responses. Zempe was already given an answer to the posed question.
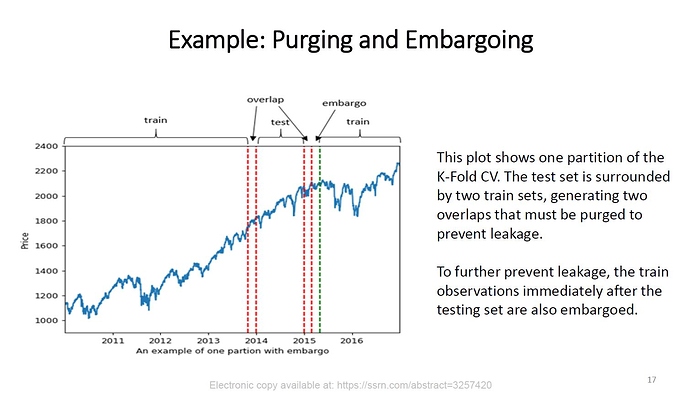
Hi @mdo. In Advances in Financial Machine Learning Marcos Lopez de Prado addresses the failures of CV for financial time series mostly because of false assumptions of observations drawn from an IID process and the leakage / overlap that results from it. As a solution Marcos suggests "purging " and “embargoing” data between training and test sets with a “Purged/Embargoed K-Fold CV”. Now in the case of applying your Era-wise Time-series CV on Numerai training data (especially Nomi) and considering that the eras provided are in chronological order would it make sense to apply such purging / embargoing? To illustrate here is a slide from Marcos’ lecture at Cornell University available online:
There really shouldn’t be a problem with overlap that purging is designed to fix, but you could always try it for added safety. I had considered adding it, but didn’t seem worth it so haven’t explored at all so ymmv. Should be pretty easy to modify my code (or sklearn’s group k-fold) to add the option.
So there is no overlap/leakage in the provided dataset. That answers my main question. Thank you @mdo
Using scipy.stats.spearmanr.correlation to score my model gives the following validation mean: 0.0250.
However, the numerai website calculates this same model having validation mean: 0.0254.
Is this small difference expected? The model is the example model: XGBRegressor(max_depth=5, learning_rate=0.01, n_estimators=2000, n_jobs=-1, colsample_bytree=0.1)
Sorry if this is a dumb question. I am a data science noob.
Did you calculate the mean of era-wise correlations?
If you just calculated the correlation ignoring the eras, that could have been the cause of the small difference?
@shonumerai123 You’re right, I just calculated the correlation ignoring the eras. My mistake, thank you for pointing this out.
So, I just went back to my code and calculated the mean of the era-wise correlations using spearmanr.correlation, here are the results:
numer.ai era-wise validation correlation: 0.0254
spearmanr era-wise validation correlation: 0.0255
So, there appears to still be a small difference!
Code snippet:
from scipy.stats import spearmanr
import numpy as np
# Numerai correlation functions
def correlation(targets, predictions):
ranked_preds = predictions.rank(pct=True, method="first")
return np.corrcoef(ranked_preds, targets)[0, 1]
def score(df):
return correlation(df[TARGET_NAME], df[PREDICTION_NAME])
#Spearman scoring
def spearmandf(df):
return spearmanr(df[PREDICTION_NAME], df[TARGET_NAME]).correlation
#read data
training_data = pd.read_csv("https://numerai-public-datasets.s3-us-west-2.amazonaws.com/latest_numerai_training_data.csv.xz")
tournament_data = pd.read_csv("https://numerai-public-datasets.s3-us-west-2.amazonaws.com/latest_numerai_tournament_data.csv.xz")
validation_data = tournament_data[tournament_data.data_type == "validation"].copy()
#features and eras
feature_names = [
f for f in training_data.columns if f.startswith("feature")
]
training_data["erano"] = training_data.era.str.slice(3).astype(int)
eras = training_data.erano
target = "target"
#model
model = XGBRegressor(max_depth=5, learning_rate=0.01, n_estimators=2000, n_jobs=-1, colsample_bytree=0.1)
model.fit(training_data[feature_names], training_data[TARGET_NAME])
#predictions
valpredictions = model.predict(validation_data[feature_names])
validation_data[PREDICTION_NAME] = valpredictions.copy()
#numerai score
validation_correlations = validation_data.groupby("era").apply(score)
print(validation_correlations.mean())
#spearman score
validation_correlations = validation_data.groupby("era").apply(spearmandf)
print(validation_correlations.mean())Yes you should use the numeral correlation function, instead of spearmanr as that is the correct scoring function. I sometimes just use spearmanr because it’s convenient and, as you found, gives virtually identical answers. The difference is spearmanr ranks both inputs (rather than just predictions) before calling the Pearson correlation function.
Here’s a slightly modified version introducing also the purging. In my understanding the embargo makes sense only for KFold CV so simply purging the periods between the Train and Test sets should be enough to avoid leakages in case of time series CV.
I haven’t had the chance to test it yet but I’ll post an update when I have run the tests.
import numpy as np
from sklearn.model_selection._split import _BaseKFold, indexable, _num_samples
class PurgedTimeSeriesSplitGroups(_BaseKFold):
def __init__(self, n_splits=5, purge_groups=0):
super().__init__(n_splits, shuffle=False, random_state=None)
self.purge_groups = purge_groups
def split(self, X, y=None, groups=None):
X, y, groups = indexable(X, y, groups)
n_samples = _num_samples(X)
n_folds = self.n_splits + 1
group_list = np.unique(groups)
n_groups = len(group_list)
if n_folds + self.purge_groups > n_groups:
raise ValueError((f"Cannot have number of folds plus purged groups "
f"={n_folds+self.purge_groups} greater than the "
f"number of groups: {n_groups}."))
indices = np.arange(n_samples)
test_size = ((n_groups-self.purge_groups) // n_folds)
test_starts = [n_groups-test_size*c for c in range(n_folds-1, 0, -1)]
for test_start in test_starts:
yield (indices[groups.isin(group_list[:test_start-self.purge_groups])],
indices[groups.isin(group_list[test_start:test_start + test_size])])Hi @mdo and others -
This is a cool way to ensure that eras/groups aren’t split across the train/test border, but I don’t see it say anywhere in the cross_val_score() docs that group info is taken into account for scoring. If it’s not, then that means the scorer is doing a spearman on the whole test set instead of averaging it over the eras. Seeing as we score on single eras in the tourney, then to get a more tournament-accurate score from cross_val_score() we need it to return the mean spearman over the various testing eras. Does anyone know how to accomplish this with sklearn’s routines?
Thanks
Even after removing eras on the border, I’m receiving significantly better results on all subsequent folds (0.05ish) vs the base fold where train=train and validation=validation (0.025ish). Is this a sign of a bug in my code, or is there something about the training vs validation set that makes the training data easier to predict in general?
I would guess, the training and validation data are selected based on their properties. For example the validation data seems to be harder than real data. Atleast in the old dataset it is the case.
For those using both neutralization and cross-validation, do you apply neutralization to the predictions of each of your folds, or only at the end after averaging the fold predictions? I think it works out to the same in the end, but this does impact the cross validation score that we use to tune hyperparameters and I don’t have a strong intuition on whether this would be desirable or not.