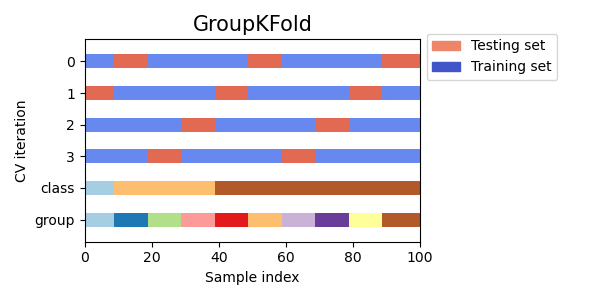
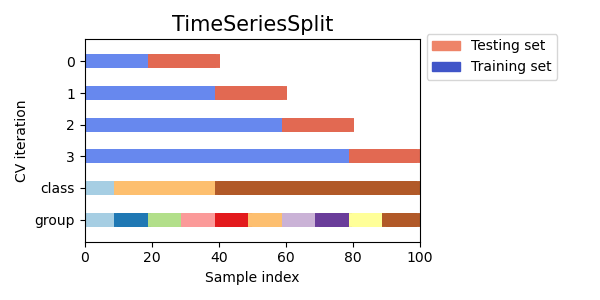
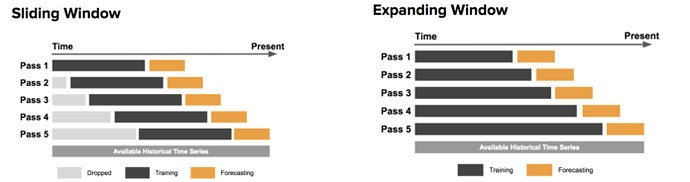
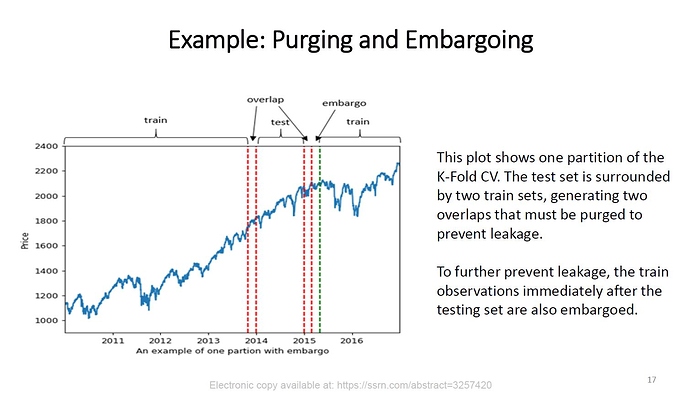
In case you’re not aware, the time-series cross-validation code in sklearn takes a groups argument, but doesn’t actually use it! I like using time-series cross-validation since it prevents you from using any future information to predict out of sample, since your out of sample test set is always in the future. I wrote a sklearn compatible cross validation splitter that can use eras as groups so your splits are always erawise. Below is example code for doing a hyperparameter grid search with XGBoost and era-wise time-series cross validation. My models Niam, NMRO, and MDO were trained in exactly this way (but with different parameter ranges than are used below). MDO also drops some of the worst and best features (according to feature importance) with the exact choices of what to drop determined by this cross validation strategy. See, nothing fancy needed to get a top 3 model  Now take this information and make even better models!
Now take this information and make even better models!
from sklearn.model_selection._split import _BaseKFold, indexable, _num_samples
from sklearn import model_selection, metrics
import pandas as pd
import numpy as np
from xgboost import XGBRegressor
import csv
from scipy.stats import spearmanr
with open('numerai_training_data.csv', 'r') as f:
column_names = next(csv.reader(f))
dtypes = {x: np.float32 for x in column_names if
x.startswith(('feature', 'target'))}
data = pd.read_csv('numerai_training_data.csv', dtype=dtypes, header=0, index_col=0)
features = [f for f in data.columns if f.startswith("feature")]
target = "target_kazutsugi"
data["erano"] = data.era.str.slice(3).astype(int)
eras = data.erano
class TimeSeriesSplitGroups(_BaseKFold):
def __init__(self, n_splits=5):
super().__init__(n_splits, shuffle=False, random_state=None)
def split(self, X, y=None, groups=None):
X, y, groups = indexable(X, y, groups)
n_samples = _num_samples(X)
n_splits = self.n_splits
n_folds = n_splits + 1
group_list = np.unique(groups)
n_groups = len(group_list)
if n_folds > n_groups:
raise ValueError(
("Cannot have number of folds ={0} greater"
" than the number of samples: {1}.").format(n_folds,
n_groups))
indices = np.arange(n_samples)
test_size = (n_groups // n_folds)
test_starts = range(test_size + n_groups % n_folds,
n_groups, test_size)
test_starts = list(test_starts)[::-1]
for test_start in test_starts:
yield (indices[groups.isin(group_list[:test_start])],
indices[groups.isin(group_list[test_start:test_start + test_size])])
def spearman(y_true, y_pred):
return spearmanr(y_pred, y_true).correlation
cv_score = []
models = []
for lr in [0.006, 0.008, 0.01, 0.012, 0.014]:
for cs in [0.06, 0.08, 0.1, 0.12, 0.14]:
for md in [4, 5, 6]:
models.append(XGBRegressor(colsample_bytree=cs, learning_rate=lr, n_estimators=2000, max_depth=md, nthread=8))
for model in models:
score = np.mean(model_selection.cross_val_score(
model,
data[features],
data[target],
cv=TimeSeriesSplitGroups(5),
n_jobs=1,
groups=eras,
scoring=metrics.make_scorer(spearman, greater_is_better=True)))
cv_score.append(score)
print(cv_score)